Abstract
Many of the available biomedical ontologies are rich in human understandable labels, but are often less rich in axiomatic descriptions. Thus, their effectiveness for supporting advanced data analysis processes is limited. Our past work included the presentation of a method for the analysis of lexical regularities in biomedical ontology labels. We showed that biomedical ontologies can present a high degree of regularity in their labels; this regularity can be used for the application of automatic axiomatic enrichment, which is the process of using textual content of the ontology to create new relationships between classes. The Gene Ontology (GO) is an ontology that is widely used for the functional annotation of genes and proteins. The class labels of the GO have a high regularity. Recently, the GO Consortium enriched the ontology by using the so-called cross-products extensions (CPE). Cross-products are generated by establishing axioms that relate a given GO class with classes from GO or other biomedical ontologies. In this paper, we study how our method for lexical analysis can identify and reconstruct the cross-products defined by the Gene Ontology Consortium. Our results show that our method is able to partially detect the GO cross-products extensions with an average recall of 61% and in some scenarios over 70%.
What could you find in this web page?
In this web page you could find the files with information about the lexical analysis and the calculation of the CPE. We have used this information to create the tables and figures used in the paper.
Source Ontology Files
- Ontology files: owl ontology files used for doing the experiment. This files were originally downloaded in OBO format. We removed the obsolete classes and transform them to the OWL format. We have in total 5 ontology files that play the role of Source Ontology or Enriching Ontology.
- Source Ontologies:
- Enriching Ontologies:
- Mungall Source Cross Products files enriched ad-hoc files from Mungall. You can load them in Protege to explore the enriched classes. If you want to see the annotations, first you need to load the original full ontologies of the cross-product.
- bpXcl (biological_process_xp_cell.obo)
- ccXcl (cellular_component_xp_cell.obo)
- goXchebi (GO_to_ChEBI.obo)
- mfXchebi (molecular_function_xp_chebi.obo)
The lexical analysis and cross-products: using the CPE metric
- Lexical Regularities and the CPE metric:
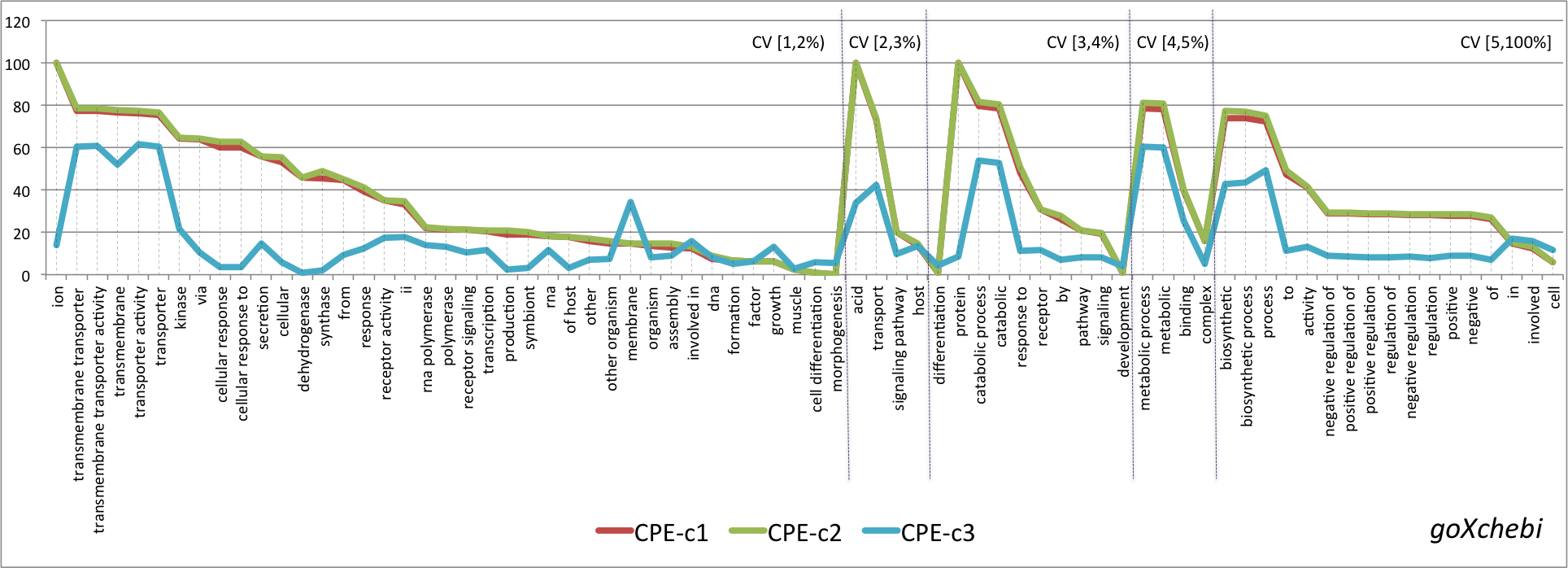
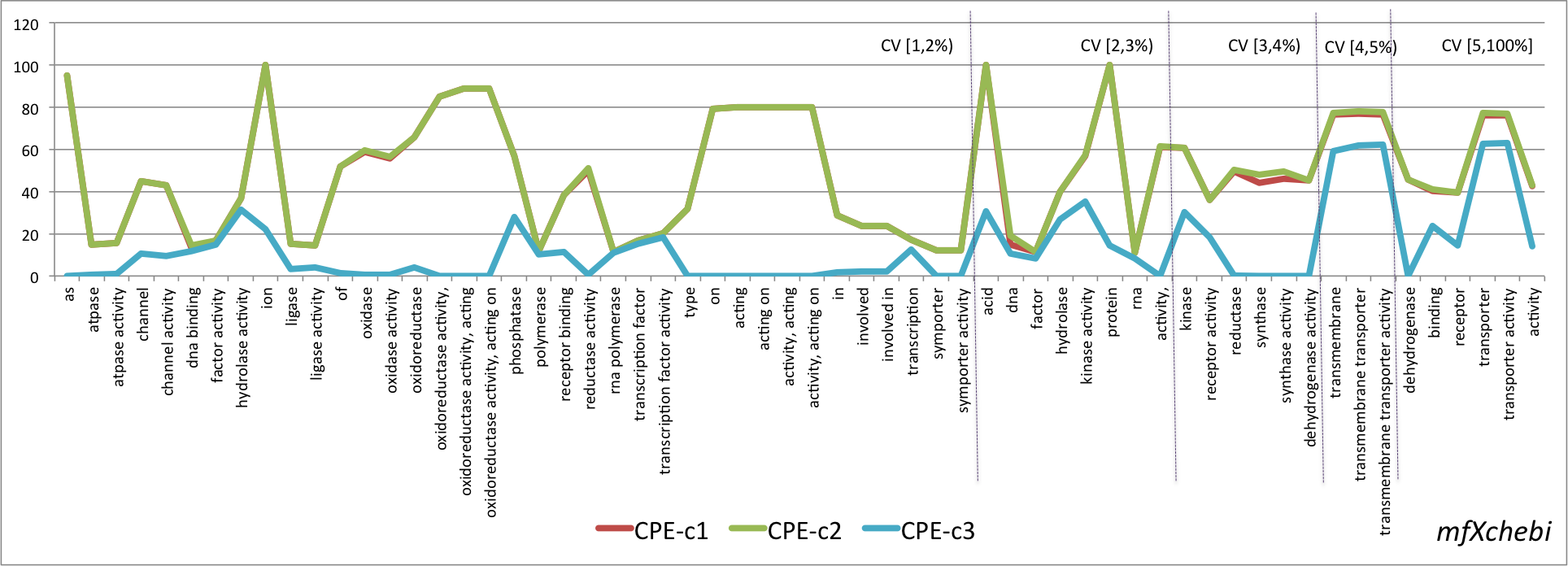
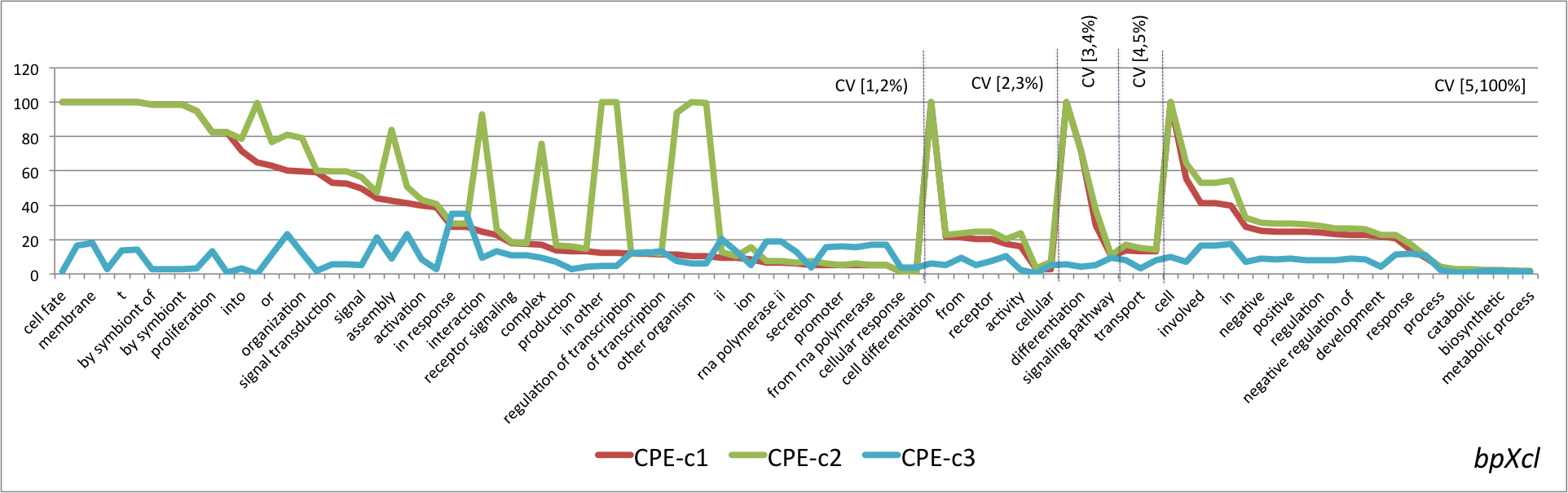
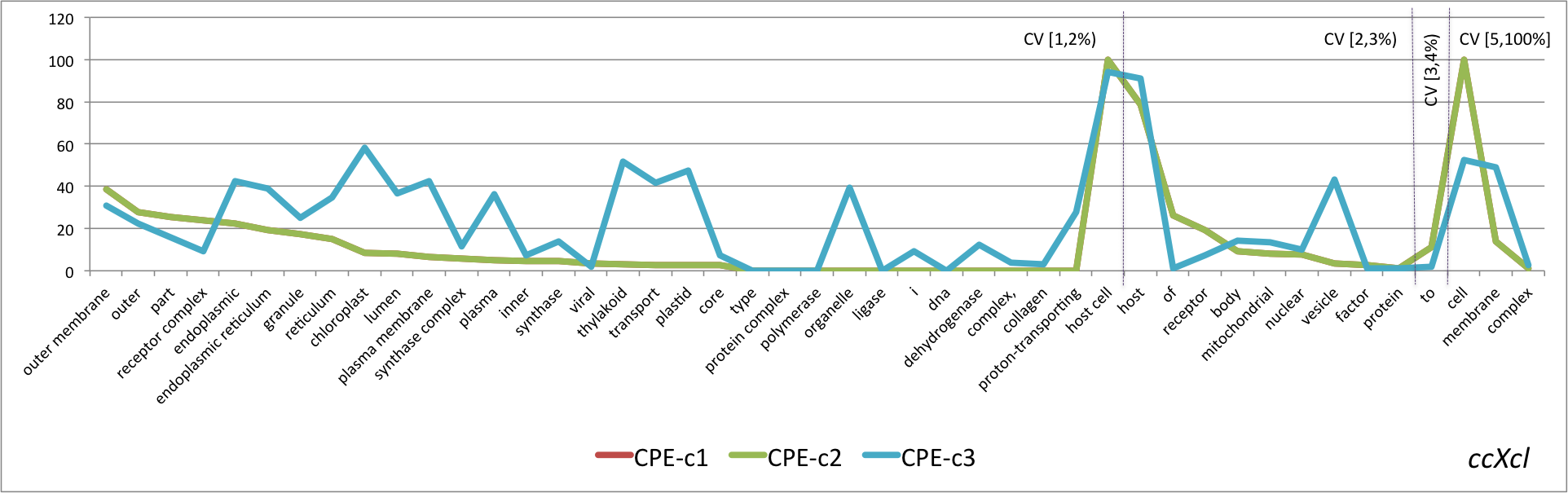
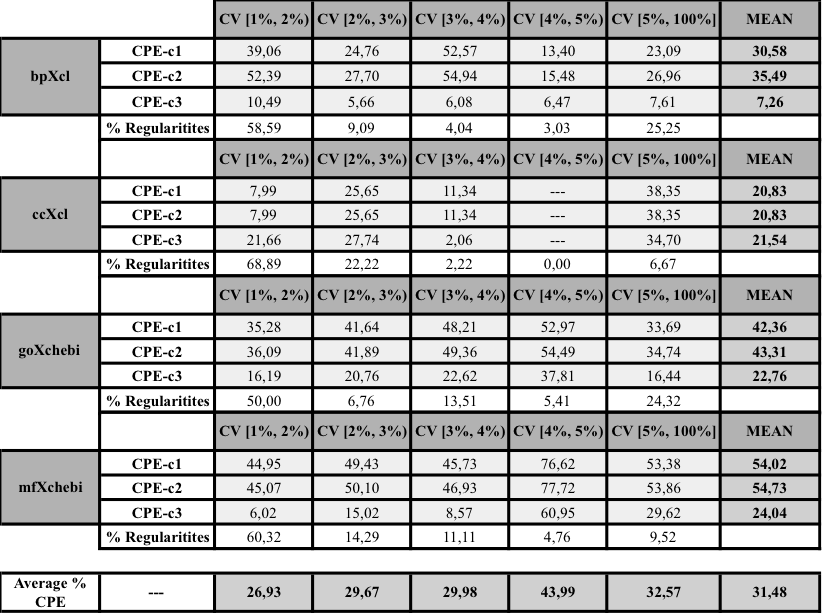
- Graphical representation of the percentage of CPE obtained for the lexical regularities found in the four pairs OSxOE. The three series represent the results for each version of the CPE. The lexical regularities are ordered first, by the coverage interval and second, by CPE-c1 value.
goXchebi
mfXchebi
bpXcl
ccXcl
- Mean values of the three versions of the CPE metric grouped by OSxOE, and by five intervals of coverage threshold.
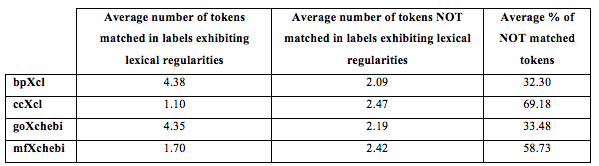
- Tokens not matched in OE: explore Excel Files with the analysis of the tokens when CPE-c3 is calculated
Comparing the CPE metric with a reference method:
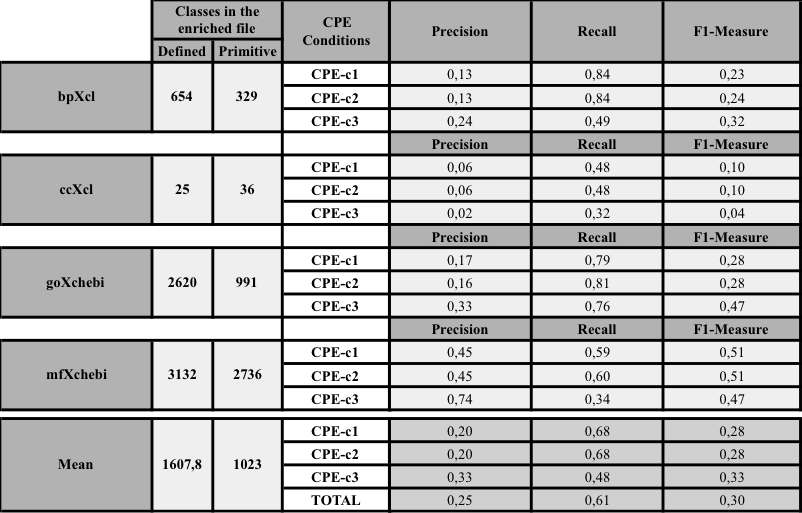
- Precision, recall and F1-Measure obtained in the comparison of our method and the reference method (Mungall et al. 2011)
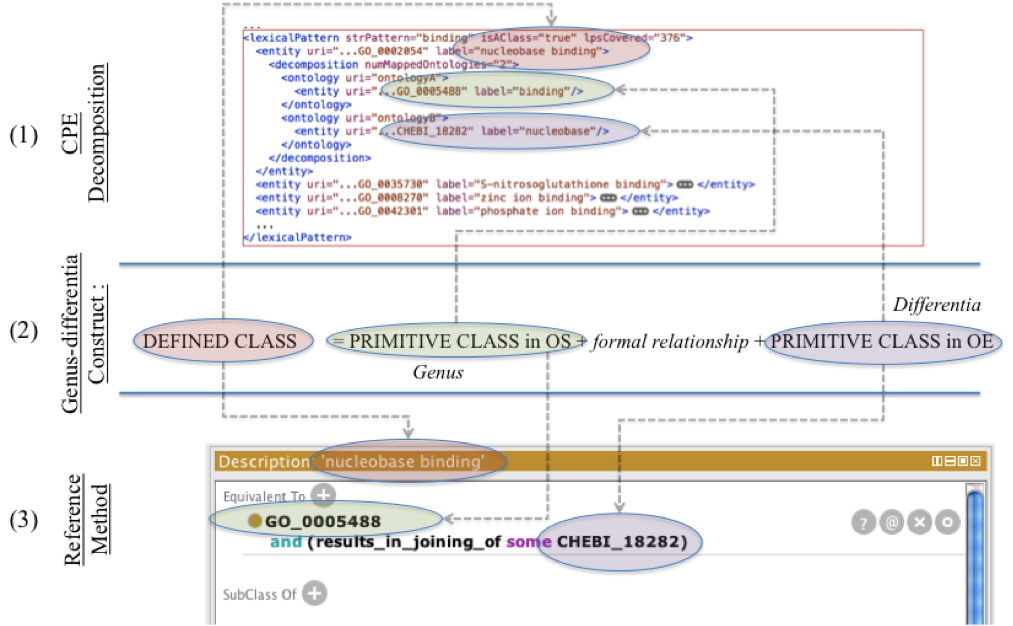
- Comparison strategy:enrichment template and equivalent elements between a CPE lexical analysis and the reference method
- XML files with the CPE decomposition of each Lexical Analysis: there is one three files for each combination of OSxOE, each file corresponds to one version of the CPE metrics.
- bpXcl
- CPE-c1 (lexAnal_bp_CaseSensitivefalse_PerctCov1.0_Reduced_by_CPE_c1.xml)
- CPE-c2 (lexAnal_bp_CaseSensitivefalse_PerctCov1.0_Reduced_by_CPE_c2.xml)
- CPE-c3 (lexAnal_bp_CaseSensitivefalse_PerctCov1.0_Reduced_by_CPE_c3.xml)
- ccXcl
- CPE-c1 (lexAnal_cc_CaseSensitivefalse_PerctCov1.0_Reduced_by_CPE_c1.xml)
- CPE-c2 (lexAnal_cc_CaseSensitivefalse_PerctCov1.0_Reduced_by_CPE_c2.xml)
- CPE-c3 (lexAnal_cc_CaseSensitivefalse_PerctCov1.0_Reduced_by_CPE_c3.xml)
- goXchebi
- CPE-c1 (lexAnal_go_CaseSensitivefalse_PerctCov1.0_Reduced_by_CPE_c1.xml)
- CPE-c2 (lexAnal_go_CaseSensitivefalse_PerctCov1.0_Reduced_by_CPE_c2.xml)
- CPE-c3 (lexAnal_go_CaseSensitivefalse_PerctCov1.0_Reduced_by_CPE_c3.xml)
- mfXchebi
- Mungall Source Cross Products files just with the Entities not detected by our method: we have removed from previous files those classes detected by our CPE metric. As we have three versions of the CPE metric, we have a reduced file for each of them.
- bpXcl
- ccXcl
- CPE-c1 (cellular_component_xp_cell.owl)
- CPE-c2 (cellular_component_xp_cell.owl)
- CPE-c3 (cellular_component_xp_cell.owl)
- goXchebi
- CPE-c1 (GO_to_ChEBI.owl)
- CPE-c2 (GO_to_ChEBI.owl)
- CPE-c3 (GO_to_ChEBI.owl)
- mfXchebi
- CPE-c1 (molecular_function_xp_chebi.owl)
- CPE-c2 (molecular_function_xp_chebi.owl)
- CPE-c3 (molecular_function_xp_chebi.owl)
- Reduced Lexical Analysis files just with information about the Entities not detected by MungalMethod method: we include in the XML of a lexical analysis reduced by those entities with CPE true if classes are found in Mungall Method or not.
Additional Files...
- Lexical Analysis Files: XML files with the result of the lexical analysis done for each Ontology Source. Independent files can be found for each combination of Source Ontology and the input parameter coverage threshold.
- bp.owl (go to folder)
- cc.owl (go to folder)
- go.owl (go to folder)
- mf.owl (go to folder)
- Explore Excel Files with CPE information and Lexical Regularitites for all the cross-products: every file conatin one sheet for each pair OSxOE